|
:: Visual presentation for circRNAs :: |
 |
|---|---|---|
The Laboratory of X-omics |
circVIS – User Manual
| This document will briefly guide user how to use the circVIS service and interpret the results. To access circVIS, please go to http://lab-x-omics.nchu.edu.tw/circVIS/. From the main page of the circVIS, an input form is provided for entering a valid gene symbol: |
|
| The default gene symbol ‘PTP4A2’ is provided as an example. At the bottom, 3 links – circVIS, HELP and Bug Report are provided. ‘circVIS’ will link to the main page, ‘HELP’ will link to this user manual and ‘Bug Report’ will link to the following page: |
|
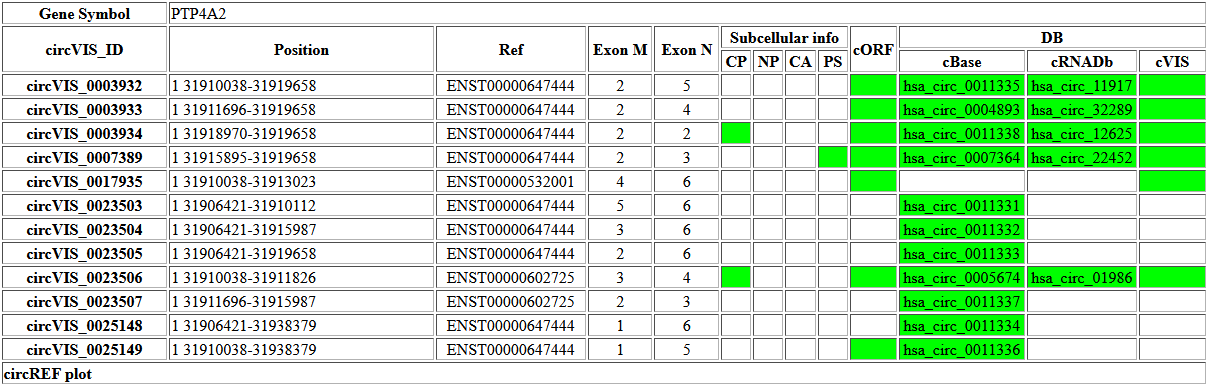
By clicking the button ‘Query’, the result page will be retrieved as following: The first part of the results is a table listing the circRNAs associated with the given gene. The origin of each circRNA is listed as chr# followed by the beginning and end site in the column of position. The column Ref and exon M/N show the backsplice taking place between the pair of exons from the representative transcript. The column of subcellular info shows ‘green’ if a given circRNA is identified in corresponding different subcellular compartments (CP: cytoplasm; NP: nucleoplasm; CA: chromatin-associated; PS: polyribosome). Similarly, if having predicted ORF, the column of ‘cORF’ (circORF) shows green. The last three columns indicate in which database (DB) the given circRNA was archived (cBase: circBase; cRNADb: circRNADb; cVIS: circVIS). |
|
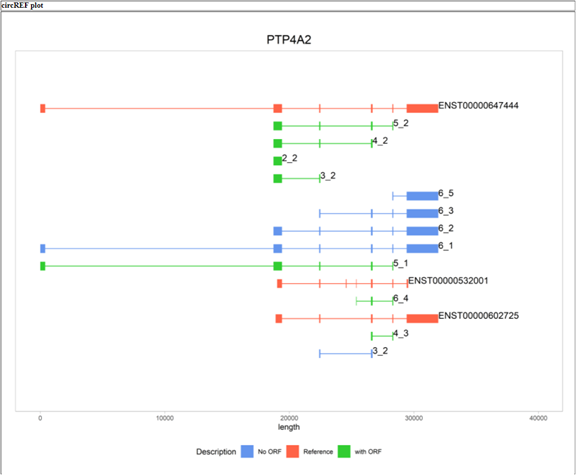
| The second part of the results shows the representative transcript(s) labeled with ENST ID (red) and circRNAs with the paired exons of backsplice (for example, ‘5_2’ stands for backsplice taking place between exon 2 and 5). In addition, circRNAs with predicted ORF are colored in green; otherwise in blue. |
|
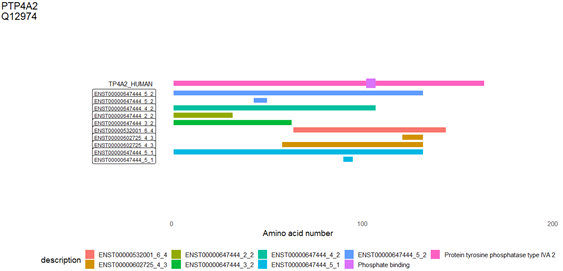
| The third part of results is a plot for mapping potential ORF to the parental protein. The gene symbol and uniprot accession number are shown at the top-left corner. The annotation features from uniprot are drawn in the first row (begin with TP4A2_HUMAN). The figure legend shown what features are. For example, PTP4A2 have two features ‘Protein tyrosine phosphatase type IVA 2’ (in pink) and ‘Phosphate binding’ (in purple). The ORFs from circRNA are labeled with corresponding representative transcript and the paired exons for backsplice (ENST_N_M). |
|
Common error messages |
|
Solution: a valid official gene symbol is required. Please go get an official gene symbol at databases such as HGNC, Ensembl and GeneCards. |
|
Cause: This means there is no circRNA identified from this gene in circBase, circRNADb or circVIS. If you think this information might not be accurate, please report it through ‘Bug report’. |
|
Cause: It is a very rare event typically caused by dis-placed output files or slow internet connection. If you think this information might not be accurate, please report it through ‘Bug report’. |
| circVIS | HELP <PDF> | Bug Report |
 Any comments can be submitted here.
Any comments can be submitted here.